PrionsPrions are infectious proteins. They constitute a fascinating class of infectious pathogens that do not rely on nucleic acids to propagate. The first prion to be discovered, PrPSc, causes fatal neurodegenerative diseases, such as Creutzfeldt-Jakob disease in humans, bovine spongiform encephalopathy (BSE, "mad cow disease") in cattle or chronic wasting disease (CWD) in cervids. During propagation of prions, what propagates is not the primary structure of the protein (as when the DNA sequence is copied); rather, what propagates is the conformation (i.e., secondary, teritary and quaternary structures) of the prion.
|
our Research
There are two main lines of research in the lab aimed at:
- Elucidating the structure of PrPSc prions in order to understand all aspects of how they propagate at the molecular level
- Identifying a therapeutic strategy to treat human prion disease
Recent results:
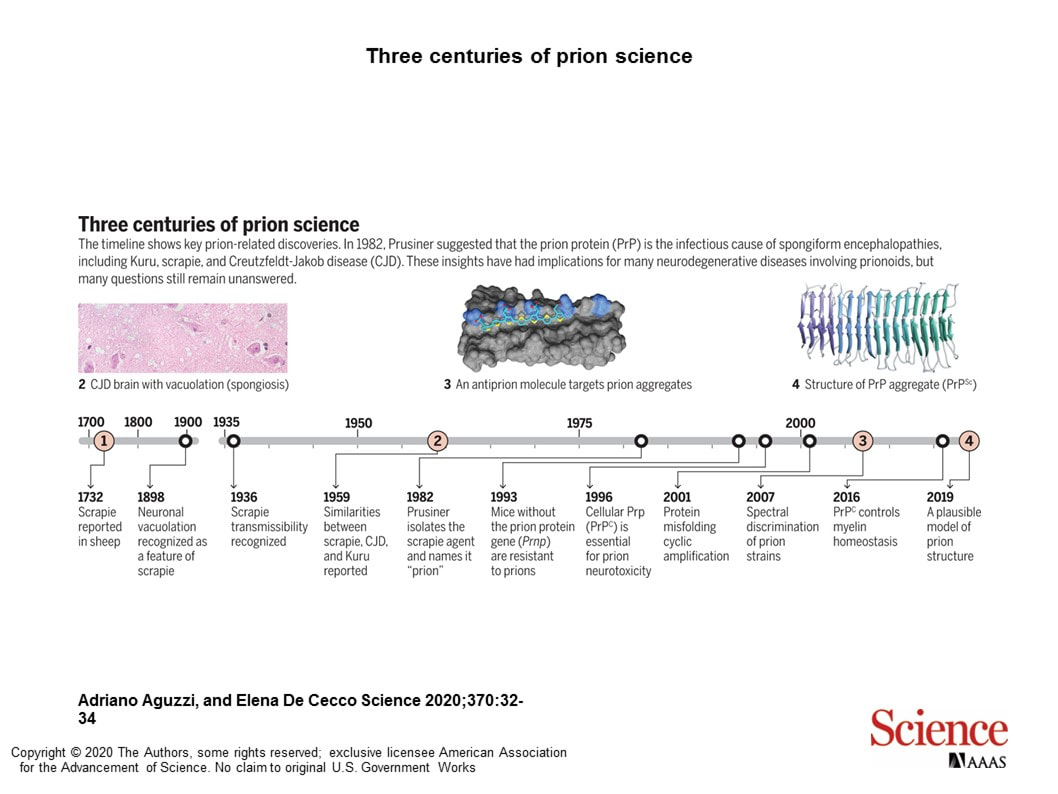
Our physically plausible PrPSc prion model chosen as one of 11 key milestones in prion research by a Perspectives article in Science (https://science.sciencemag.org/content/370/6512/32).
Haz clic aquí para editar.
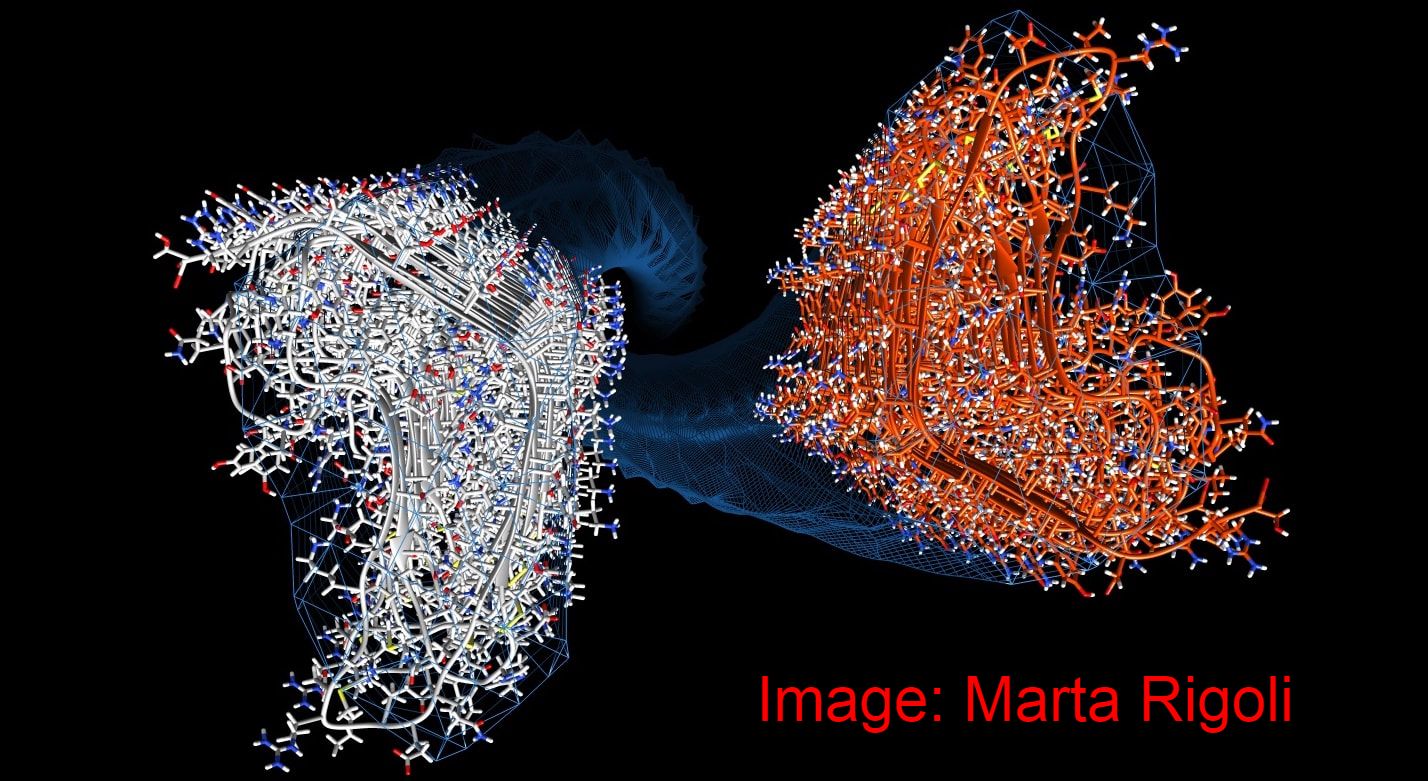
A physically plausible atomistic model of PrPSc generated:
In collaboration with Giovanni Spagnolli, Marta Rigolli, Pietro Facciolli, Emiliano Biasini (University of Trento) and other colleagues, we have generated the first physically plausible, stable, atomistic model of PrPSc. The model, based on a 4-rung beta solenoid, complies with all the available structural constraints derived from cryo-EM, FTIR, limited proteolysis etc. Based on the model, we have also modelled for the first time the PrPSc-templated conversion of PrPc to PrPSc. The stdy has been published in PLoS Pathogens:
https://journals.plos.org/plospathogens/article/comments?id=10.1371/journal.ppat.1007864
See the PrPC to PrPSc conversion movie here:
doi.org/10.1371/journal.ppat.1007864.s009
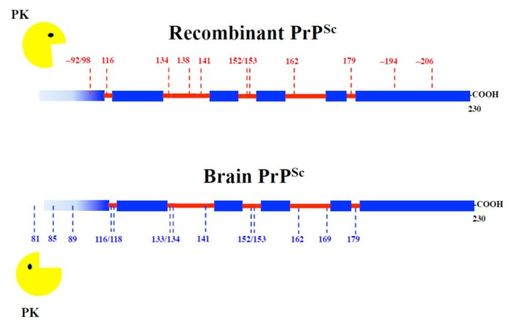
Recombinant PrPSc and brain-derived PrPSc
share a similar architecture:
Recombinant PrPSc prepared in our lab following a "recipe" developed by Joaquín Castilla and cols. (CIC-BIOGUNE, Bilbao, Spain) has essentially the same architecture as PrPSc isolated from the brain of prion-infected animals. We compared patterns of limited proteolysis using epitope mapping and mass spectrometry to identify proteinase K-resistant fragments. These results pave the way to use recombinant PrPSc for NMR-based structural studies, currently ongoing in our lab.
(Sevillano et al., 2018, PLoS Pathog 14(1): e1006797).
share a similar architecture:
Recombinant PrPSc prepared in our lab following a "recipe" developed by Joaquín Castilla and cols. (CIC-BIOGUNE, Bilbao, Spain) has essentially the same architecture as PrPSc isolated from the brain of prion-infected animals. We compared patterns of limited proteolysis using epitope mapping and mass spectrometry to identify proteinase K-resistant fragments. These results pave the way to use recombinant PrPSc for NMR-based structural studies, currently ongoing in our lab.
(Sevillano et al., 2018, PLoS Pathog 14(1): e1006797).
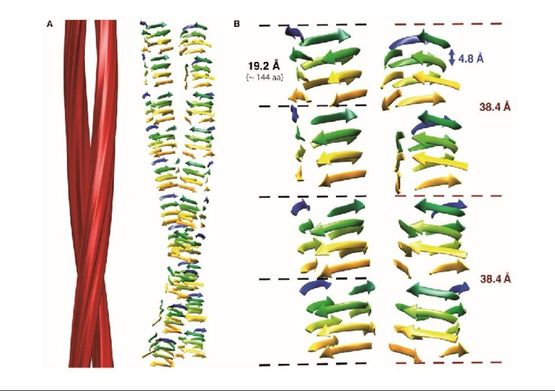
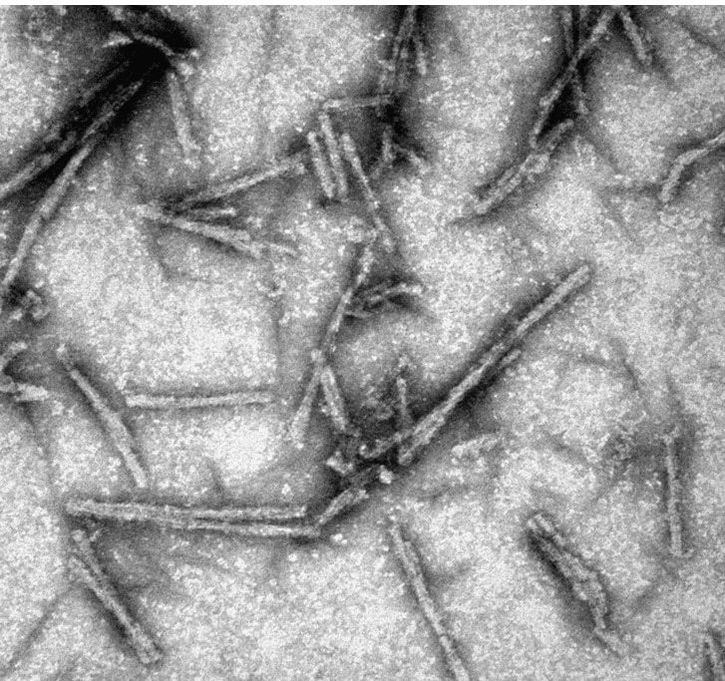
The structural architecture of PrPSc revealed using cryo-electron microscopy
In collaboration with Matthijn Vos, from the FEI Company, we obtained cryo-EM images of unprecedented quality of GPI-anchorless PrPSc prion fibers. Fourier transform of images showed that even individual beta strands stacked along the fiber axes are discerned. Then, in collaboration with Holger Wille and Howard Young, from the University of Alberta, we carried out sophisticated image analysis techniques to reconstruct the fibers. Our results showed that PrPSc prion fibers are made up of two intertwined protofilaments, each one composed of stacked PrPSc subunits, and that each PrPSc subunit must be a 4-rung beta solenoid (Vázquez-Fernández et al., 2016, PLoS Pathog. 12(9):e1005835). This structural knowledge has allowed for the first time to understand the mechanism of propagation of PrPSc prions:
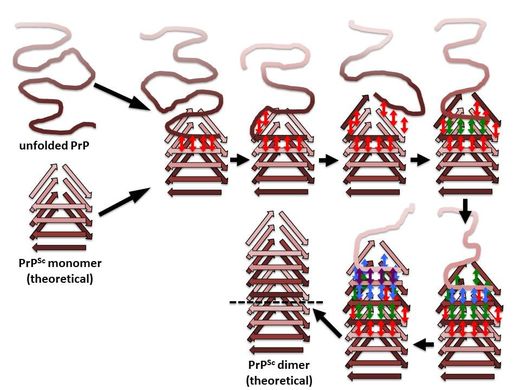
An incoming molecule of unfolded PrP would interact with an uncapped β-solenoid surface and adopt a β-strand conformation by forming backbone hydrogen-bonds (red arrows). Once the first rung of a nascent β-solenoid configuration has been formed, it would self-template successive rungs of β-solenoid structure using the same mechanism (green, blue, and purple arrows, respectively). Once the fourth and final rung has been templated, a new molecule of PrPSc is formed and the original template surface has been re-created;from: Wille & Requena, 2018, Pathogens 7(1) E20.
NEWS
Leticia Camila Fernández Flores, new Master ´ s graduate from the Prionlab, July 2020.
Nuria López Lorenzo and Yaiza B. Codeseira resenting their work at the 2019 CIMUS Workshop.
Join the lab
We are currently accepting applications to join the lab for Ph.D. studies. We are also willing to accept Erasmus + and other visiting researchers for short research stays.
Si tienes un buen expediente y te interesa unirte al laboratorio para un TFG, TFM o tesis doctoral, contacta con:
[email protected]
Requena´s lab Christmas lunch 2019